Demo 8: Riemann Integrals in multiple Dimensions and Variable Substitution#
Demo by Christian Mikkelstrup, Hans Henrik Hermansen, Jakob Lemvig, Karl Johan Måstrup Kristensen, and Magnus Troen
from sympy import *
from dtumathtools import *
init_printing()
Python Functions for Parametrizations#
When we work with change of variables, Python and lambda functions cna be useful for handling Sympy objects. To illustrate how this can be applied we will consider the example
We would like to determine \(f(\boldsymbol{r}(u,v,w))\).
For this, we now define \(f\) as, respectively, a Sympy expression (variable), a standard Python function, and a lambda function.
x,y,z = symbols('x,y,z', real =True)
u,v,w = symbols('u,v,w', real =True)
# The parameter function
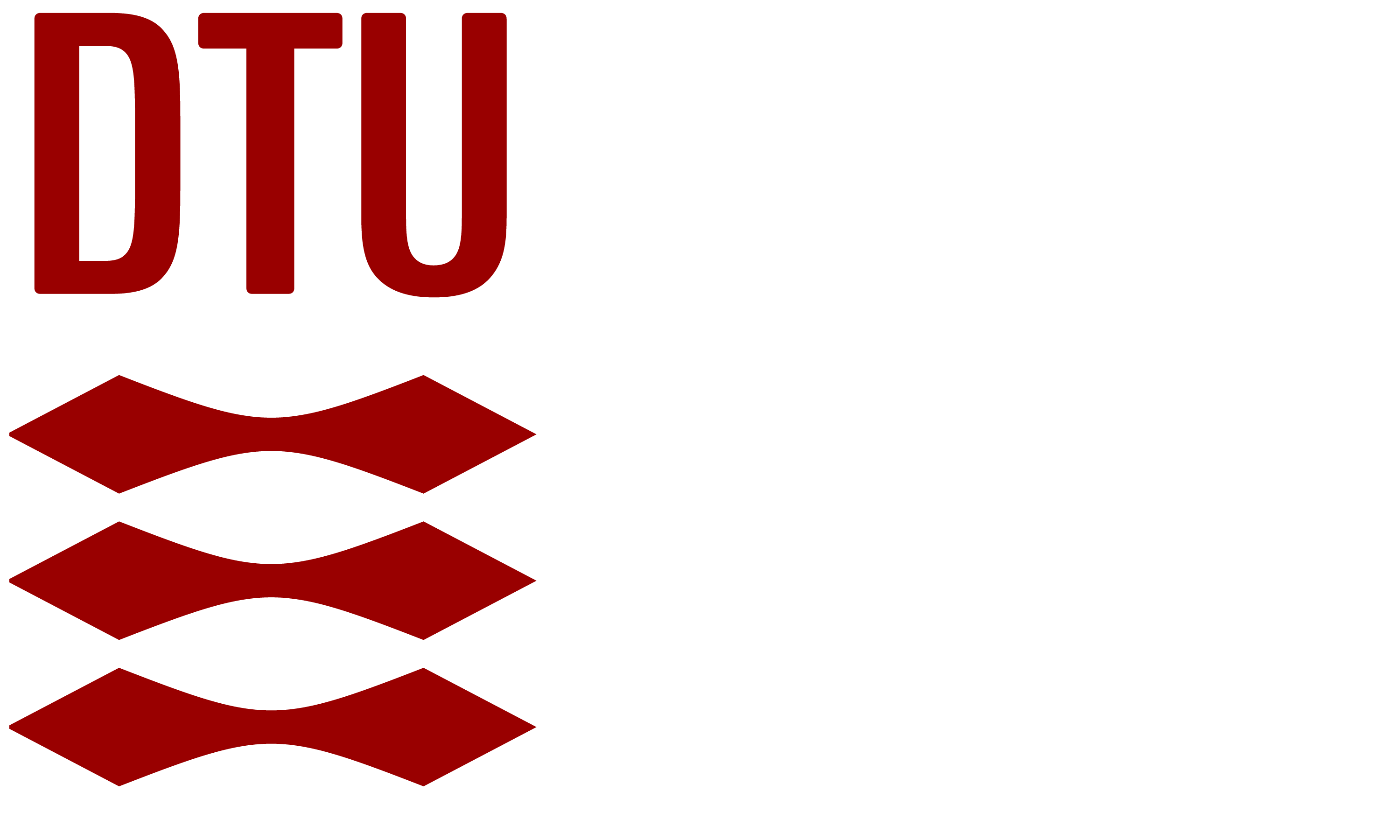
r = Matrix([u -v, w**2, v*(u+w)])
# Functions
f_sym = x+y+z
def f_function(x,y,z):
return x+y+z
f_lam = lambda x,y,z : x+y+z
As a Sympy expression, \(f(\boldsymbol{r}(u,v,w))\) can be written using
fr_sym = f_sym.subs(zip((x,y,z), r)).simplify()
With the Python and lambda functions we could alternatively simply do
fr_function = f_function(*r).simplify()
fr_lam = f_lam(*r).simplify()
This does though require that one remembers *! This tells Python that the coordinates in \(\boldsymbol{r}\) are to be given to the function as individual arguments.
fr_sym, fr_function, fr_lam
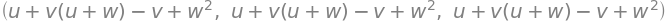
Integration of Regions Boudned by Straight Lines#
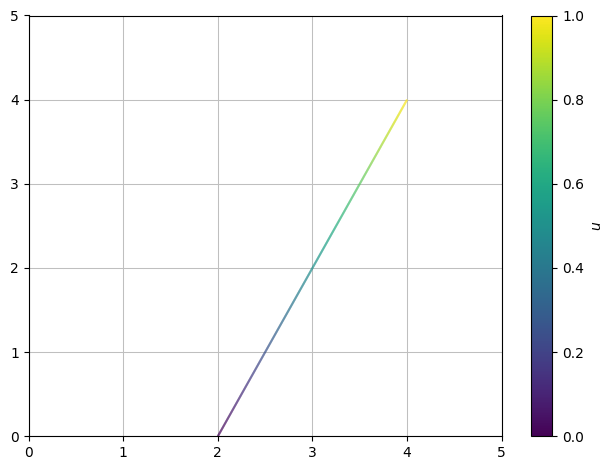
The simplest parameter curve is a straight line. An example of this is the line between the points \((2,0)\) and \((4,4)\):
parametrized by
where \(u \in [0,1]\). When the parameter \(u\) runs through the interval \([0,1]\), the point \((x,y)\) will travel along the line starting at \((2,0)\) and ending at \((4,4)\).
u,v = symbols("u v")
r_L = Matrix([2,0]) + Matrix([2,4]) * u
r_L
dtuplot.plot_parametric(*r_L, (u,0,1), xlim=(0,5), ylim=(0,5)) # The * symbol in front of r_l is important to remember when one plots parametric curves
<spb.backends.matplotlib.matplotlib.MatplotlibBackend at 0x7fca5b04e670>
We can often simplify our lives a bit when we are to parametrize planar regions by considering line segment. Let us have a look at two examples:
Example#
We are to parametrize the region in the plane that is bounded by the lines:
\(y = 1-x\)
\(y = 2x+1\)
\(x=2\)
which written out as a set of points is
Let us see how this region looks:
x,y = symbols("x y")
region = dtuplot.plot_implicit(Eq(x,2),Eq(y,2*x+1),Eq(y,1-x),(x <= 2) & (y <= 2*x+1) & (y >= 1-x),(x,-0.1,5),(y,-1.1,5.1),aspect="equal",size=(8,8))
/builds/pgcs/pg14/venv/lib/python3.9/site-packages/spb/series.py:2214: UserWarning: The provided expression contains Boolean functions. In order to plot the expression, the algorithm automatically switched to an adaptive sampling.
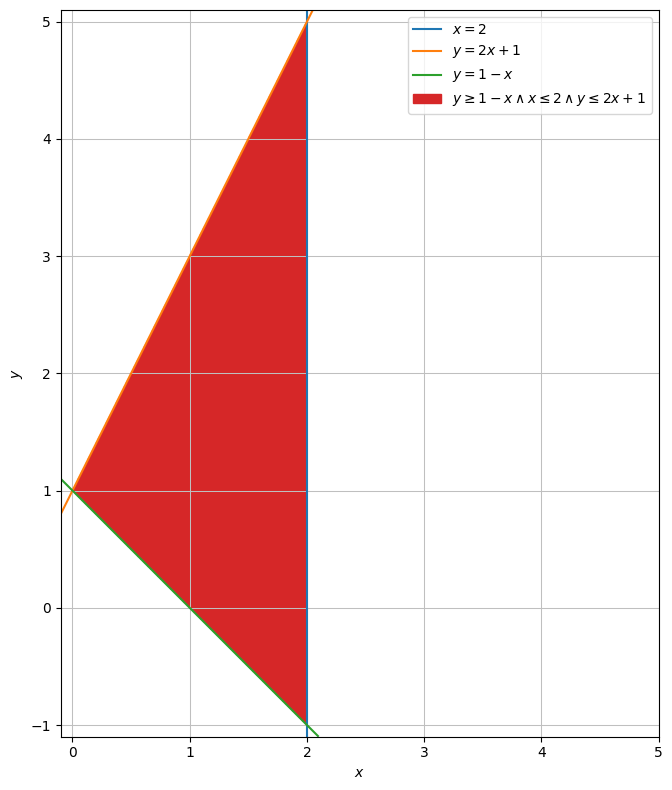
We want to parametrize the entire region. This means that all lines that can be spanned from the graph \(y=-x + 1\) to the graph \(y=2x + 1\) must be described.
We describe the points at the bottom of the arrows generally with \(A = (u,1-u)\).
The top can be described generally by \(B = (u,2u+1)\).
Thus, the directional vector \(AB\) can be described by
AB = dtuplot.quiver(Matrix([1,0]),Matrix([0,3]),show=False,rendering_kw = {"color" : "black"})
region.extend(AB)
region.show()
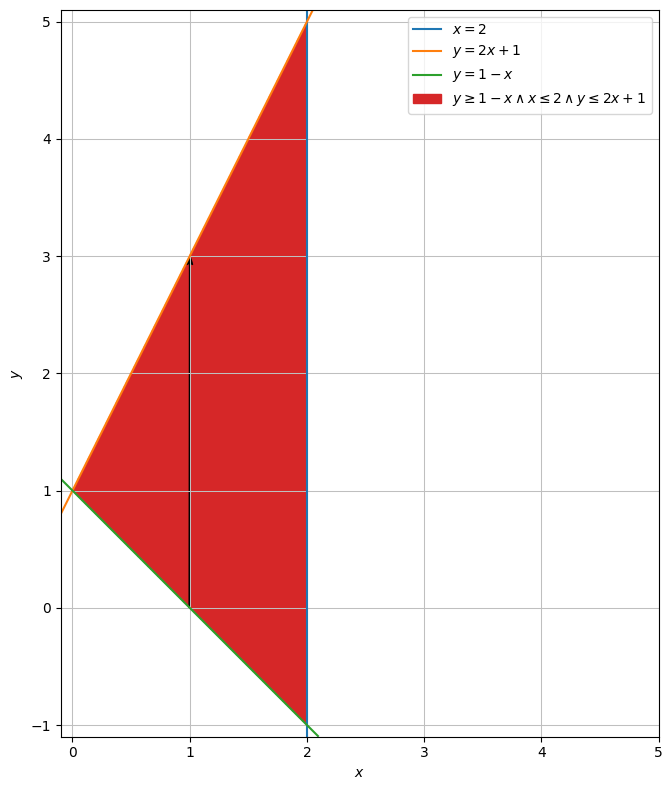
We can then parametrize a given line segment between two points on top and on the bottom in the following way:
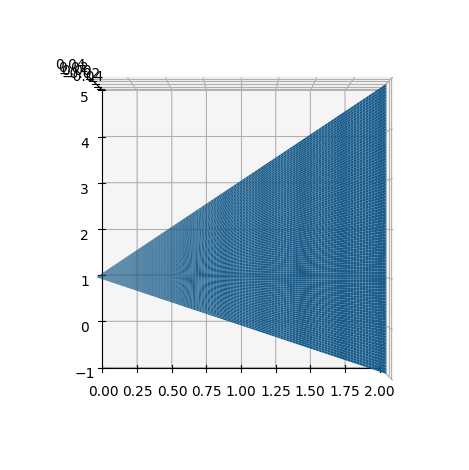
If we choose a \(u\) that runs through the interval \([0,2]\), then we want to run through all lines from left to right, and in this way cover the entire triangle. So, the entire region can be parametrized as:
dtuplot.plot3d_parametric_surface(u,1-u+3*u*v,0,(u,0,2),(v,0,1),camera={"elev":90, "azim":-90})
## One can not plot planes in 2D, so we will use the "camera" argument to look at the plot from above
/builds/pgcs/pg14/venv/lib/python3.9/site-packages/spb/backends/matplotlib/matplotlib.py:599: UserWarning: Attempting to set identical low and high zlims makes transformation singular; automatically expanding.
<spb.backends.matplotlib.matplotlib.MatplotlibBackend at 0x7fca5c1b8d90>
Regions Bounded by Circles#
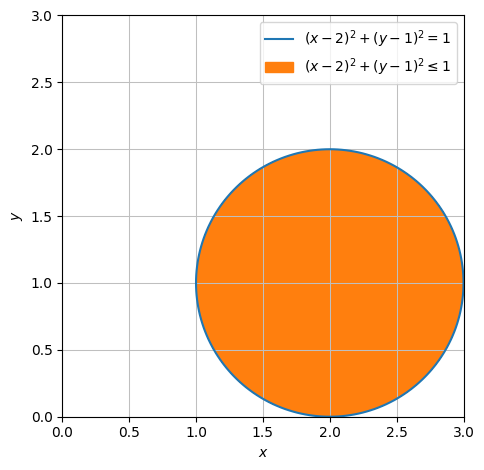
We will now parametrize the unit circle disc centred at \((2,1)\). Since we already know how a circle can look, we will directly aim towards finding the line segment that we want to parametrize.
circle = dtuplot.plot_implicit(Eq((x-2) ** 2 + (y-1) ** 2,1),((x-2) ** 2 + (y-1) ** 2 <=1),(x,0,3),(y,0,3), aspect = 'equal',show=False)
AB = dtuplot.quiver(Matrix([2,1]),Matrix([cos(pi /4),sin(pi / 4)]),rendering_kw={"color":"black"}, show=False)
# circle.extend(AB)
(circle + AB).show()
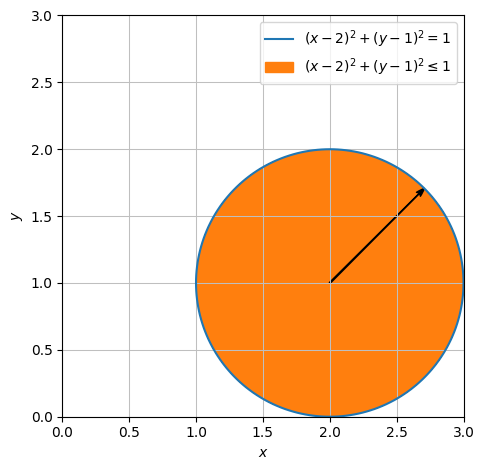
We will now parametrize all lines that stetch from the centre to a point on the circle disc boundary. This is done in the following way. Our \(A\) is now \((2,1)\), our \(B\) is \((2 + \cos(u), 1 + \sin(u))\), and so our directional vector \(AB\) is \(\begin{bmatrix} \cos(u) \\ \sin(u) \end{bmatrix}\). This specific line is thus parametrized as:
If we let \(u\) run through the interval \([0,2 \pi]\), we get all lines between the centre and the boundary, and in this way we get all of the circle disc. So, the surface is parametrized with:
rC = Matrix([2,1]) + v * Matrix([cos(u),sin(u)])
rC
dtuplot.plot_parametric_region(*rC, (u,0,2*pi), (v,0,1), aspect="equal")
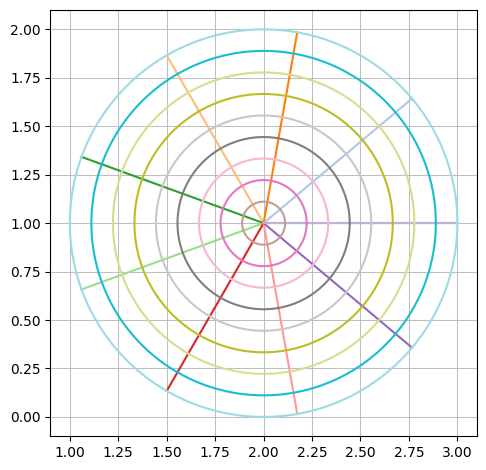
<spb.backends.matplotlib.matplotlib.MatplotlibBackend at 0x7fca38f37ca0>
For each of these regions in the plane, a plane integral for a given function can be computed.
Plane Integral#
With the function
f = lambda x,y: 2 * x * y
f(x,y)
we wish to compute the plane integral of \(f\) over the region that is bounded by our circle, so \(\int_C f(x,y)\; \mathrm{d}\pmb{x}\).
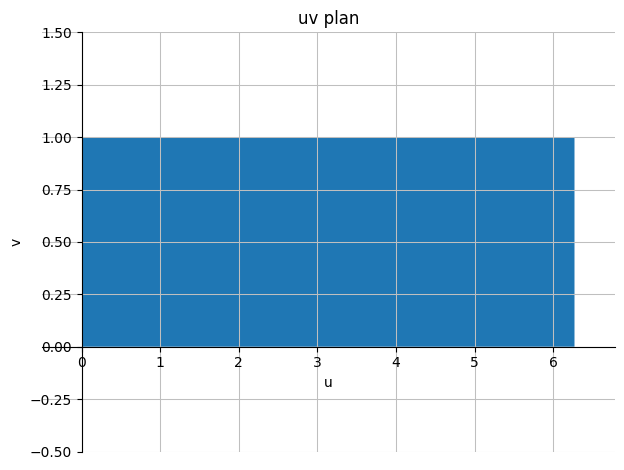
Note: With this parametric representation, the axis-parallel square \([0, 2\pi] \times [0, 1]\) in the \((u,v)\) plane is mapped to the circle we integrate over in the \((x,y)\) plane.
dtuplot.plot_implicit(((x < 2*pi) & (x > 0) & (y > 0) & (y < 1)), (x, -0.5, 2*pi + 0.5), (y, -0.5,1.5),
title="uv plan", xlabel="u", ylabel="v", axis_center='auto')
print("mapped over")
circle.show()
mapped over
Now we will need the absolute value of the Jacobian determinant, which locally measures how much the \((u,v)\) parameter region is deformed when it is undergoing the map \(\boldsymbol r_C\). And since there are multiple variables and derivatives, we first set up the Jacobian matrix:
JacobiM = Matrix.hstack(rC.diff(u), rC.diff(v))
JacobiM
Now we can find the absolute value of the Jacobian determinant \(|\det(\boldsymbol J_{\boldsymbol r_C})|\) by
Jacobian = abs(det(JacobiM)).simplify()
Jacobian

The integral of \(f\) over \(C\) is then according to the change-of-variables theorem given by:
integrate(f(*rC) * Jacobian,(u,0,2*pi),(v,0,1))
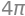
Volume Integrals: The Weight of a Sphere in \(\mathbb{R}^3\)#
Let
So, \(\Omega\) describes a unit sphere in \(\mathbb{R}^3\). Now let
describe a constant “mass density function” (so, all parts of the sphere are assumed to weight equally much). We wish to determine the mass of the sphere described by \(\Omega\), so we are to compute
This requires that we first and foremost must determine a parametrization of the sphere as well as the corresponding Jacobian determinant. The easiest would be to use spherical coordinates as described in the book, but we will try to work our way towards a parametrization based on what we know about the parametrization of a circle.
Parametrization of a Sphere#
u,v,w = symbols('u v w', real = True)
We will use the approach that a sphere can be described as a circle in the \((x,y)\) plane rotated \(180\) degrees about the \(z\) axis. The circle is parametrized as follows:
r_circle = Matrix([cos(u), 0, sin(u)])
dtuplot.plot_parametric(r_circle[0], r_circle[2], (u,0,2*pi), use_cm=False,aspect="equal", xlabel = 'x', ylabel = 'z')
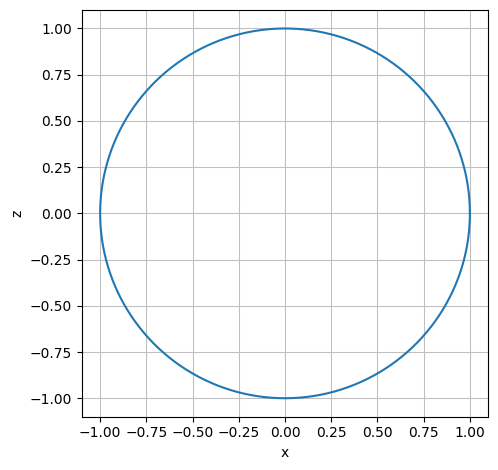
<spb.backends.matplotlib.matplotlib.MatplotlibBackend at 0x7fca390e96a0>
From here the circle can be rotated about the \(z\) axis using the rotation matrix
This will give a sphere with a radius of \(1\).
Rz = Matrix([[cos(v), -sin(v), 0], [sin(v), cos(v), 0], [0, 0, 1]])
r_sphere = simplify(Rz * r_circle)
r_sphere
Finally we can achieve a solid sphere by introducing the parameter \(w \in [0,1]\), which takes care of reaching all radii between \(0\) and \(1\). So, the final parametrization for the sphere is:
r_ball = r_sphere * w
r_ball
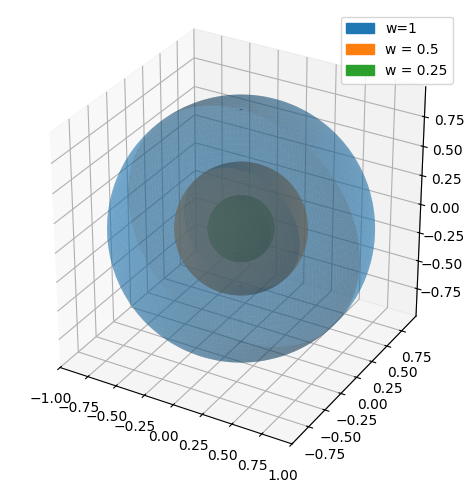
This can be visualized by looking at \(\boldsymbol{r}_{ball}\) with different values of \(w\):
big_ball = dtuplot.plot3d_parametric_surface(*r_ball.subs(w,1), (u, 0, 2*pi), (v, 0, pi), aspect = 'equal', label = 'w=1', rendering_kw = {'alpha': 0.4,}, show = false)
half_ball = dtuplot.plot3d_parametric_surface(*r_ball.subs(w,0.5), (u, 0, 2*pi), (v, 0, pi),label ='w = 0.5', rendering_kw = {'alpha': 0.5}, show = False)
quarter_ball = dtuplot.plot3d_parametric_surface(*r_ball.subs(w,0.25), (u, 0, 2*pi), (v, 0, pi),label ='w = 0.25', show = False)
(big_ball + half_ball + quarter_ball).show()
The Jacobian Determinant and Density Function#
The Jacobian determinant \(\det(\boldsymbol{J}_{\boldsymbol r_{ball}})\) is found by
This can easily be done with dtumathtools, where we include the absolute value:
J_ball = r_ball.jacobian([u,v,w])
J_ball
Jacobian = abs(det(J_ball)).simplify()
Jacobian

The density function in this case is the \(1\)-function and is thus not necessary to include, but let us define it for the sake of formality:
f = lambda x,y,z: 1
Determining the Integral#
Now we can compute the mass of the sphere by
First we define and simplify the integrand:
integrand = (f(*r_ball) * Jacobian).simplify()
integrand

M = integrate(integrand, (u, 0, 2*pi), (v, 0, pi), (w, 0, 1))
M
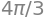
Note that \(M\) is equal to the volume of a sphere with a radius of \(1\), which makes senes since all points have been assigned with mass density of \(1\).
Another Density Function#
Now, let
describe a new density function.
Let us compute
f1 = lambda x,y,z: 2*sqrt(x**2 + y**2 + z**2)
integrand = (f1(*r_ball) * Jacobian).simplify()
integrand

M1 = integrate(integrand, (u, 0, 2*pi), (v, 0, pi), (w, 0, 1))
M1