Demo 9: Line and Surface Integrals of Scalar Functions#
Demo by Christian Mikkelstrup, Hans Henrik Hermansen, Jakob Lemvig, Karl Johan Måstrup Kristensen, and Magnus Troen
from sympy import *
from dtumathtools import*
init_printing()
x,y,z = symbols('x y z', real=True)
Curve Lengths#
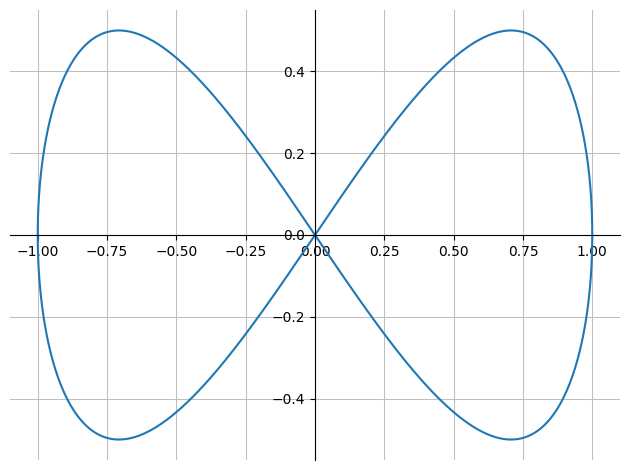
We are given a parameter curve,
u,v = symbols('u v', real=True)
r = Matrix([sin(u), sin(u)*cos(u)])
r
where \(u \in [0, 2\pi]\).
p_curve = dtuplot.plot_parametric(*r, (u,0,2*pi), use_cm=False, label="r(u)",axis_center="auto")
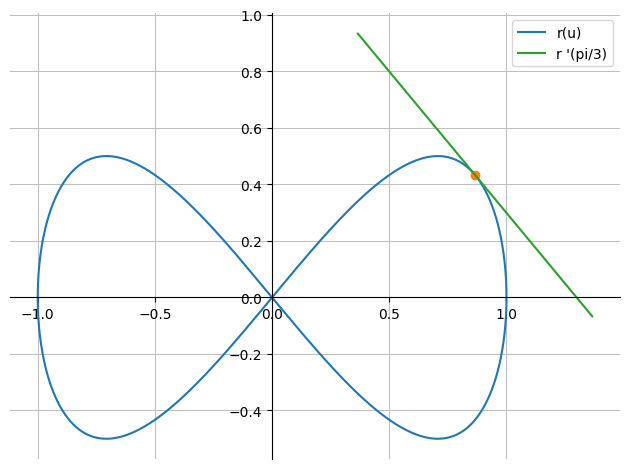
Tangent Vector and Tangent#
We find the tangent vector,
dr = r.diff(u)
dr
We now find the parametric representation for the tangent corresponding to the curve point \(r(\pi/3)\),
t = symbols("t")
r_tan = r.subs(u,pi/3) + t*dr.subs(u,pi/3)
r_tan
p_point = dtuplot.scatter(r.subs(u,pi/3), show=False)
p_tan = dtuplot.plot_parametric(*r_tan, (t,-1,1), use_cm=False, label="r '(pi/3)", show=False)
(p_curve + p_point + p_tan).show()
Length of the Curve#
And then the length of this curve can be found by
Jacobian = dtutools.l2_norm(dr)
integrate(Jacobian, (u,0,2*pi)).n()
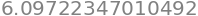
Line Integral in 3D space#
We are given a function:
x,y,z = symbols("x y z")
f = lambda x,y,z: sqrt(x**2 + y**2 + z**2)
f(x,y,z)

and a parameter curve (a so-called space curve)
r = Matrix([u*cos(u), u*sin(u), u])
r
for \(u\in[0,5]\).
p_spacecurve = dtuplot.plot3d_parametric_line(*r, (u,0,2*pi), use_cm=False, label="r(u)",aspect="equal", legend=True)
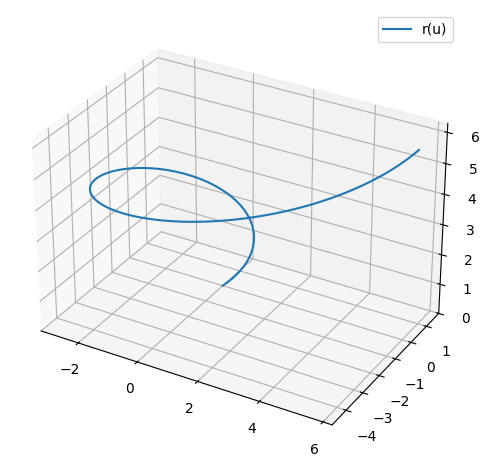
The restriction of the function to the curve is:
restriction = f(*r).simplify()
restriction

and if one remembers that \(u\) is positive, since we have defined \(u\in [0, 5]\), then the absolute value is irrelevant. From the definitions of our u and v we do though have them defined by
u,v = symbols('u v', real=True)
where Sympy only takes into account the assumption that \(u=|u|\) in exactly this case. If we had defined them using
u,v = symbols('u v', real=True, nonnegative=True)
instead, we could have used refine() and Q.\textit{assumption}(symbol), where assumption can be replaced by the entries in this table.
We shall here use Q.nonnegative(), and then Sympy shows that the restriction in fact is
true_restriction = refine(restriction, Q.nonnegative(u)) # Q.nonnegative(u) tells refine() that u >= 0
true_restriction

for \(u \in [0,5]\). Whether we write \(u\) or \(|u|\) in the expression can sometimes make a difference if Sympy tries to integrate it.
Let’s return to the line integral that we wish to compute: \(\int_K f(x,y,z)\, \mathrm{d}\pmb{s}\).
First, we find the tangent vector,
dr = r.diff(u)
dr
The length of the tangent vector \(||r_u'(u)||\) is equal to the Jacobian,
Jacobian = dtutools.l2_norm(dr).simplify()
# The following lines only works if $u$ is a real variable
# meaning if 'u = symbols('u', real=True)':
# Jacobian = dr.norm()
Jacobian
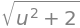
We can now find the integral along the curve,
integrate( f(*r) * Jacobian ,(u,0,5)).evalf()
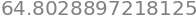
and the length of the curve,
integrate(Jacobian,(u,0,5)).evalf()

Integral over Cylinder Surface in \(\mathbb{R}^3\)#
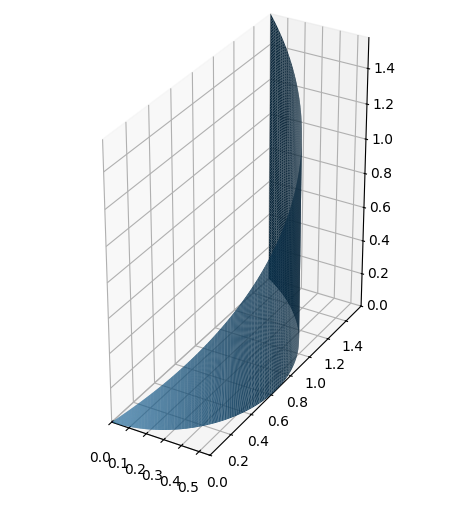
We consider a function \(f: \mathbb{R}^3 \to \mathbb{R}\) given by
We also consider a surface given by the following parametric representation with \(u \in [0,\frac{\pi}{2}]\) and \(v \in [0,1]\):
# This time we remember 'nonnegative=True', since we again see that
# none of the intervals for u and v contain negative numbers
u,v = symbols('u v', real=True, nonnegative=True)
r = Matrix([u*cos(u),u*sin(u),u*v])
def f(x,y,z):
return 8*z
r, f(x,y,z)
dtuplot.plot3d_parametric_surface(*r,(u,0,pi/2),(v,0,1), aspect='equal')
<spb.backends.matplotlib.matplotlib.MatplotlibBackend at 0x7f7faabeedc0>
Side Note: Interactive 3D plots#
Note: This is not needed. Personally, though, we think that it is nice to be able to turn and rotate 3D plots, and have a better overview over what is happening in the plot.
Second note: Apart from that, one also must be aware that one’s notebook cannot be exported to PDF, if one has plots of this type in the document. There are work-arounds for this, but do consider whether you want to spend time on figuring it out.
If one wants to be able to move a 3D plot in order to achieve a better sense of the plot, it is an option to use another backend when plotting. This does though require that one installs the package plotly with pip:
pip install plotly
Or by removing the commenting from the cell below and execute it a single time. Then plotly will be installed in the version of Python that your notebook is using right now.
# ! pip install plotly
# vvvvvvvvvvvvvvvvvv here
# dtuplot.plot3d_parametric_surface(*r,(u,0,pi/2),(v,0,1), aspect='equal', backend=dtuplot.PB, use_cm=True)
The Jacobian of a Surface in 3D#
We find the Jacobian and insert the parametric representation in \(f\):
crossproduct = r.diff(u).cross(r.diff(v))
Jacobian = sqrt((crossproduct.T * crossproduct)[0]).simplify()
Jacobian

integrand = f(*r) * Jacobian
integrand

integrate(integrand,(v,0,1),(u,0,pi/2)).evalf()